Many more African Americans (AA) die from prostate cancer than Caucasian Americans (CA), even after factoring in socioeconomic influences. The NIH designated identifying medically relevant differences in these populations as a high priority research area. Inventors at GWU identified differences in gene splicing between AA and CA prostate cancer samples. Gene splicing variation causes the functional products of a gene to include different parts in different people. The inventors identified hundreds of differentially spliced genes. Importantly, 13 of those genes contribute to oncogenic signaling pathways. One example is the oncogene PIK3CD, which has a highly aggressive AA splicing variant not found in CA prostate cancer samples. Applying knowledge of aggressive splicing variants will enable diagnostic and prognostic testing for prostate cancer. Population-specific diagnosis increases efficiency and enables delivery of personalized therapeutics.
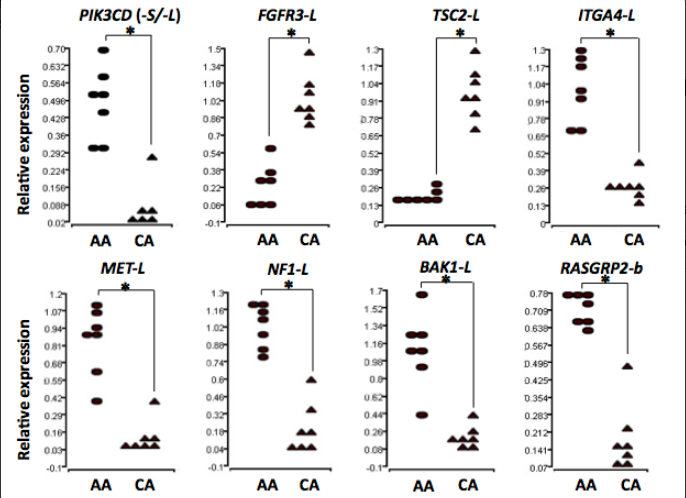
Figure: Quantitative PCR validates splice variants found in prostate cancer samples from different populations.
Applications:
- Personalized therapeutics for prostate cancer
- Diagnostics for prostate cancer
- Assays for determining aggressiveness of prostate cancer
- Assays for determining prognosis of prostate cancer patients
- Prostate cancer genetic risk determination
Advantages:
- Novel markers for prostate cancer aggressiveness
- Oncogenic splicing variants may be specifically targeted for degradation